Specialized metabolic enzymes originated from primary metabolism, yet their catalytic properties differ significantly from their counterparts in primary metabolism. Whereas primary metabolic enzymes are highly specific and exhibit low levels of mechanistic elasticity, specialized metabolic enzymes, on average, display greater catalytic promiscuity and evolvability. Comparative study of structurally related primary and specialized metabolic enzyme families offers unique opportunities to unveil determining factors of enzyme evolvability. Combining structural biology, biochemistry, computational chemistry, and artificial intelligence (AI) approaches, we aim to decipher design principles that govern important parameters regarding enzyme evolution, such as protein folding stability and catalytic properties. We aim to leverage the knowledge gained from this line of research to ultimately enable enzyme function prediction based on primary sequence, as well as de novo enzyme design.
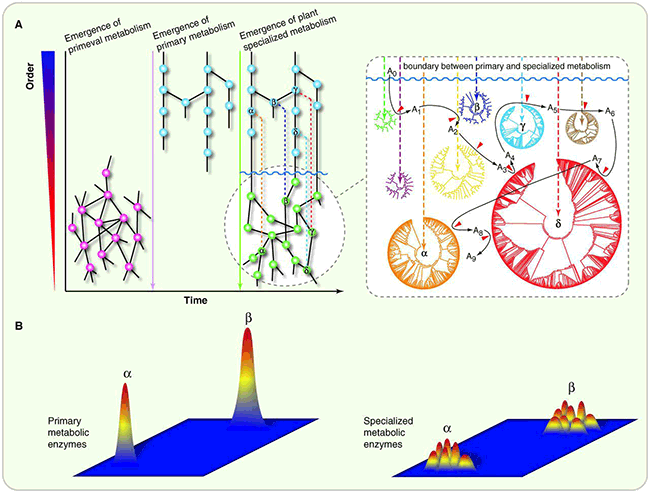
Patterns of emergence and evolution of primary and specialized metabolism. (A) Primary metabolism likely arose from promiscuous primeval metabolic reactions and evolved toward greater catalytic precision and efficiency. Specialized metabolism likely emerged from primary metabolism. Due to early gene-duplication events, the functional constraints acquired by primary metabolic enzymes were released, allowing the mutational exploration of new areas of enzyme chemistry. Enzymes and reactions are represented by nodes (pink, blue, and green spheres) and links (black lines), respectively. The right panel illustrates the stepwise assembly of a specialized metabolic pathway using descendents from enzyme folds rooted in primary metabolism (indicated by circular phylogenetic trees and highlighted with Greek letters). Products of one reaction serve as substrates for another. Red arrowheads indicate the recruitment of single enzymes from protein families. (B) Hypothetical catalytic landscapes of primary and specialized metabolic enzymes relating sequence variation (horizontal plane) to the breadth of disparate enzymatic activities of stable protein folds (vertical axis). Catalytic specificity and efficiency for primary metabolic enzymes are maintained by natural selection, constraining their chemical mechanisms. Specialized metabolic enzymes often produce additional products from a single enzyme due to expanded substrate recognition and/or multiple chemical transformations within a single enzyme.

