Random mutations are generally thought to have deleterious effects on protein folding and function. Although proteins are marginally stable and exhibit a level of mutational robustness, they do misfold under excessive environmental stresses or mutations. In human, continual aggregation of certain misfolded proteins could lead to pathology, including many neurodegenerative diseases such as Alzheimer’s, Huntington (HD), Parkinson’s, and the prion maladies. In addition to proteins, it was recently discovered that RNA in a cell is also susceptible to aggregation. Base-pairing mediated clustering of RNA contributes to the pathology of nucleotide repeat expansion diseases such as HD, myotonic dystrophy, and certain forms of ALS. Defects in RNA metabolism are frequently observed in many neurodegenerative disorders, and circumstantial evidence (such as mislocalization of RNA binding proteins, RNA accumulation in tau and Aβ aggregates suggests that RNA aggregation could be commonplace in degenerative disorders. Indeed, recent studies show that the prevalence of RNA aggregation is likely underestimated due to technical difficulties in probing aggregated RNA.
Plants contain rapidly evolving specialized metabolic system, and presumably encounter destabilized evolutionary intermediates along their mutational trajectories. Have plants evolved unique molecular mechanisms that assist folding of those destabilized proteins and RNAs and or mitigate cytotoxicity arising from protein and RNA aggregation? We use the plants as a unique system to examine the in vivo function and behavior of aggregation-prone model proteins and RNAs as well as mutant enzymes that exhibit broadened product promiscuity and or decreased folding stability. We attempt to identify genetic components and specialized metabolites involved in mitigating cellular stress caused by protein and RNA aggregation in plants.
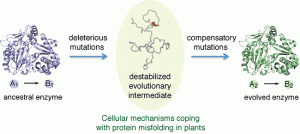
Protein misfolding stress is indeed an evolutionary problem. Cellular mechanisms that mitigate proteolytic stress due to misfolded proteins may also play an important role in broadening evolutionary trajectories for evolving enzymes.

